Here we introduced the concept of independent component analysis ICA to single-cell clustering and network analysis. A common example application is the cocktail party problem of listening in.

The Escherichia Coli Transcriptome Consists Of Independently Regulated Modules Biorxiv
Motivation Single cell RNA sequencing scRNA-seq enables transcriptional profiling at the level of individual cells.

Independent component analysis rna-seq. In this context a methodology that can be used to cluster small series of distribution-free data while accounting for the dependence between subsequent time-points should be used for temporal analysis RNA-seq data. See Method Details of independently acting factors or independent components ICs. ICA has been extensively applied to gene expression data since the age of microarray 22 23.
Independent Component Analysis ICA is a statistical procedure that uses a transformation to convert raw time series data into sets of values of independent variables which can be used for cluster analysis to identify sets of genes with similar temporal expression patterns. Independent Component Analysis ICA is a statistical procedure that uses a transformation to convert raw time series data into sets of values of independent variables which can be used for cluster analysis to identify sets of genes with similar temporal expression patterns. Independent component analysis ICA is a matrix factorization approach where the signals captured by each individual matrix factors are optimized to become as mutually independent.
Independent Component Analysis ICA is a statistical procedure that uses a transformation to convert raw time series data into sets of values of independent variables which can be used for cluster analysis to identify sets of genes with similar temporal expression patterns. Compatible with Illumina NGS systems. The Independent Component Analysis ICA 9 is a statistical procedure that uses a transformation to convert raw time series data into sets of values of independent variables which can be used for cluster analysis to identify sets of genes with similar temporal expression patterns.
Ad Finish rRNA depletion library generation in under 65 hours. Should be used for temporal analysis RNA-seq data. Coli transcriptome as determined by Independent Component Analysis ICA is conserved across multiple independent datasets including both RNA-seq and microarray datasets.
Here the authors identify statistically independent gene sets by applying independent component analysis to a high-quality E. Each independent component was characterized by a co-expression pattern and was. We introduced independent component analysis ICA into single cell clustering to decompose the gene expression matrix into a number of independent components.
ICA is a special case of blind source separation. NIFA is a general approach advancing scRNA-seq analysis capabilities and it allows researchers to better take advantage of their data. In signal processing independent component analysis is a computational method for separating a multivariate signal into additive subcomponents.
Here we show that the underlying structure of the E. DOAJ is a community-curated online directory that indexes and provides access to high quality open access peer-reviewed journals. With the emergence of high-throughput platforms datasets comprising tens of thousands or more cells have become routine and the technology is having an impact across a wide range of biomedical subject areas.
Ad Finish rRNA depletion library generation in under 65 hours. This is done by assuming that the subcomponents are non-Gaussian signals and that they are statistically independent from each other. Gene expression time series GETS analysis aims to characterize sets of genes according to their longitudinal patterns of expression.
Compatible with Illumina NGS systems. Learn at your own pace by doing interactive coding exercises. We applied independent component analysis ICA to decompose the heterogeneity of scRNA-seq profiles into a relatively small number 30 as it was close to the estimated optimal number.
ICA has also been used to predict gene co-expression modules in bulk RNA-seq. The Independent Component Analysis ICA is a statistical procedure that uses a transformation to convert raw time series data into sets of values of independent variables which can. Learn at your own pace by doing interactive coding exercises.
Mchikinapittedu 1 Introduction Single-cell RNA. However scRNA-seq data are high-dimensional and affected by. Due to the large.
Improving Gene Function Predictions Using Independent Transcriptional Components Nature Communications

Slicer Inferring Branched Nonlinear Cellular Trajectories From Single Cell Rna Seq Data Biorxiv

Single Cell Rna Seq Of Rheumatoid Arthritis Synovial Tissue Using Low Cost Microfluidic Instrumentation Biorxiv
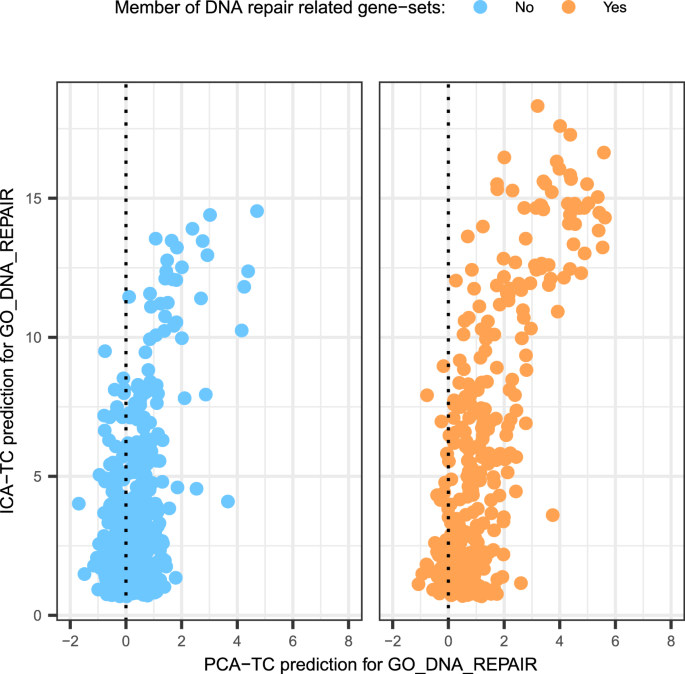
Ijms Free Full Text Independent Component Analysis For Unraveling The Complexity Of Cancer Omics Datasets Html

Independent Component Analysis Uncovers The Landscape Of The Bladder Tumor Transcriptome And Reveals Insights Into Luminal And Basal Subtypes Cell Reports
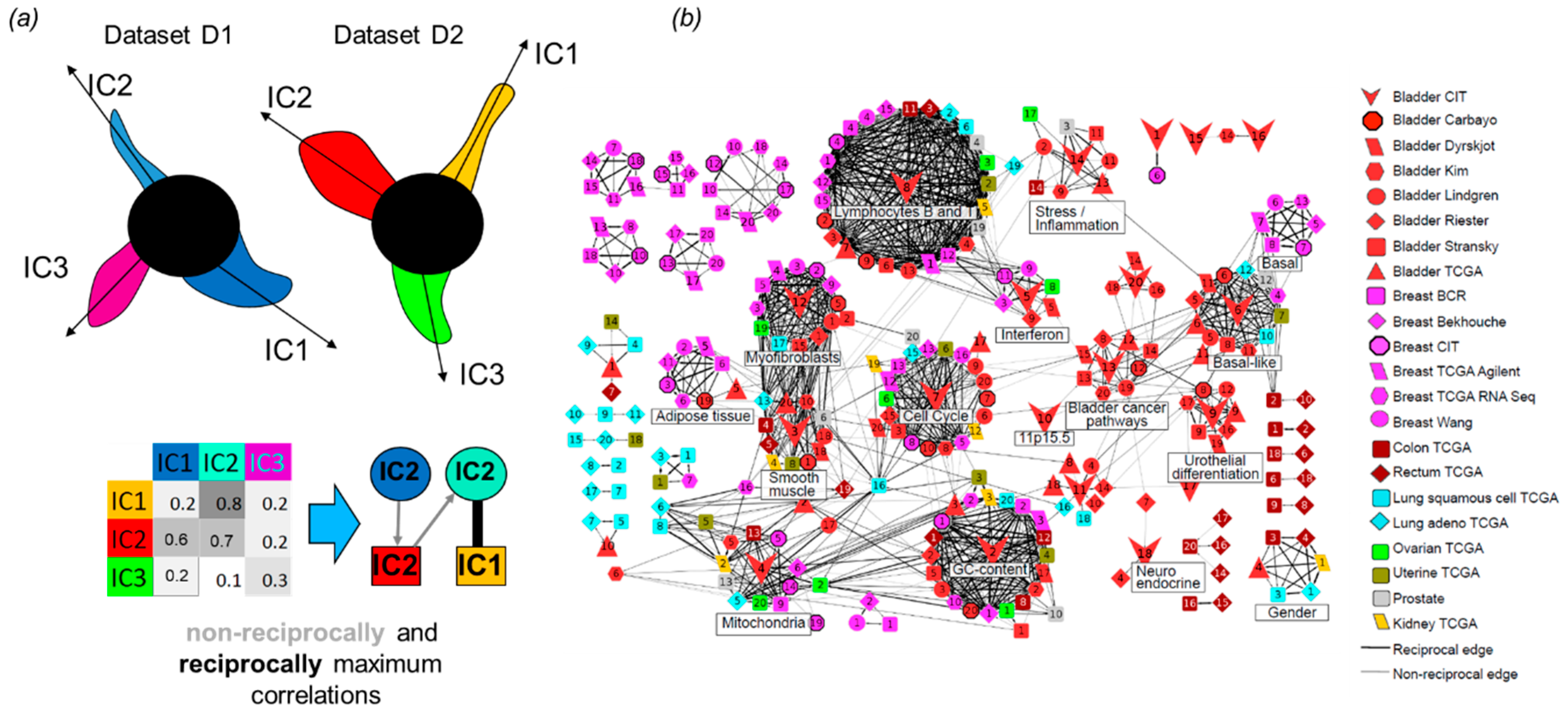
Ijms Free Full Text Independent Component Analysis For Unraveling The Complexity Of Cancer Omics Datasets Html

Single Cell Rna Seq Theory Of Normalization And Pca Introduction To Single Cell Rna Seq Archived

The Escherichia Coli Transcriptome Consists Of Independently Regulated Modules Biorxiv
Qc Methods For De Analysis Using Deseq2 Introduction To Dge Archived

The Escherichia Coli Transcriptome Consists Of Independently Regulated Modules Biorxiv

Principal Components Analysis Pca And Hierarchical Heat Map Cluster Download Scientific Diagram
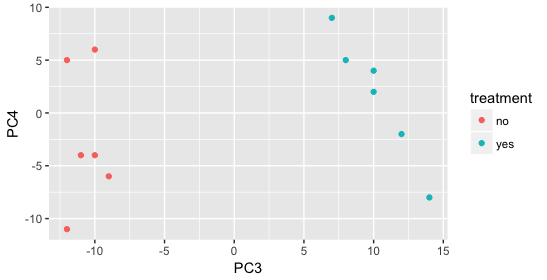
Qc Methods For De Analysis Using Deseq2 Introduction To Dge Archived
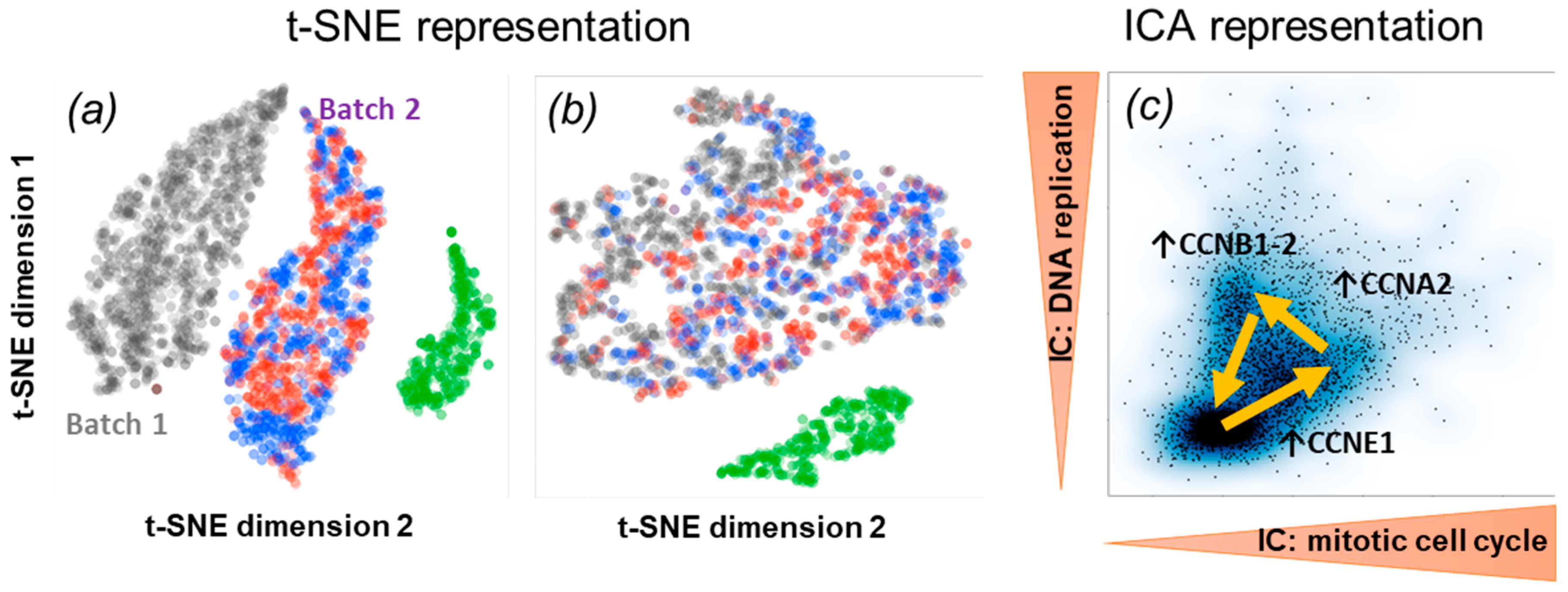
Ijms Free Full Text Independent Component Analysis For Unraveling The Complexity Of Cancer Omics Datasets Html

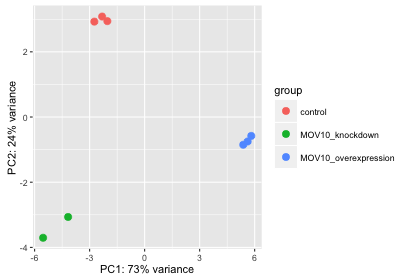
Principal Component Analysis A The Percent Variability Explained By Download Scientific Diagram

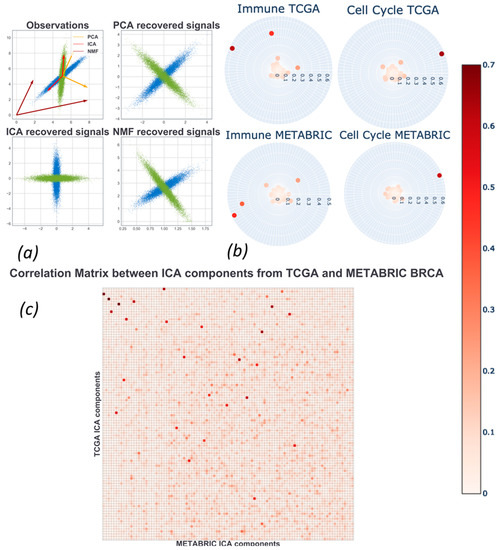
Ijms Free Full Text Independent Component Analysis For Unraveling The Complexity Of Cancer Omics Datasets Html

Independent Component Analysis Of E Coli S Transcriptome Reveals The Cellular Processes That Respond To Heterologous Gene Expression Sciencedirect

Cancers Free Full Text Mining Of Self Organizing Map Gene Expression Portraits Reveals Prognostic Stratification Of Hpv Positive Head And Neck Squamous Cell Carcinoma Html
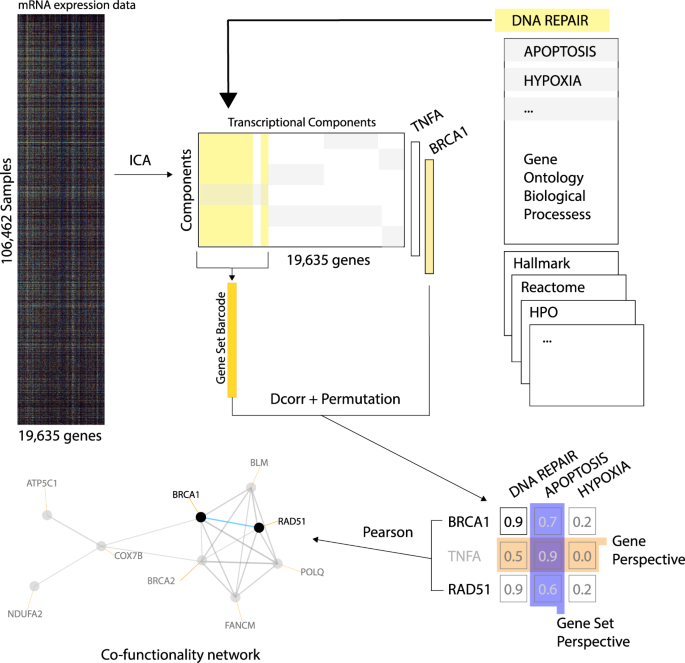
Improving Gene Function Predictions Using Independent Transcriptional Components Nature Communications

Performance Assessment And Selection Of Normalization Procedures For Single Cell Rna Seq Biorxiv

0 Comments